Discovering the Mysteries of Epilepsy Using pClamp Analysis
Summary
Epileptic seizures are rapid and rhythmic electrical bursts in
the brain that disrupt normal activity. More then 2 million Americans
have epilepsy, a disorder which can be caused by problems such as
abnormal brain development, brain damage from illness or injury
and genetic mutations.
.gif)
Inherited syndromes account for about 5% of all epilepsy cases.
In this later category is a syndrome called generalized epilepsy
with febrile (fever) seizures (GEFS+). The cause of this syndrome
seems to involve a genetic mutation of the sodium ion (Na+)
channels in certain cells. These channels are donut-like pores that
act like gates that let electrical current in the form of ions flow
into the cell and then close. The closing, or inactivation, occurs improperly in mutant channels.
In 1998, researchers at Vanderbilt and elsewhere were able to identify
the first Na+ channel gene mutation for inherited epilepsy.
Later, additional mutations were identified. The researchers then
replicated "healthy" as well as mutant versions of this Na+ channel in the laboratory to study the functional effects of the mutation at the molecular level.
The experimental data produced during the studies at Vanderbilt
consisted of normalized current measurements from excited tissues
resembling mutant and non-mutant ion channels as they were subjected
to three different pulse protocols. From the resulting data, the researchers
built an accurate model that characterized the Na+ channels.
The model allowed a clear understanding of this malfunction and
became a significant system to test drugs to restore proper function
to these channels.
How Origin Was Used
The researchers at Vanderbilt took advantage of Origin's ability
to import and plot pClamp data, as well its ability to perform
nonlinear curve fitting with the Nonlinear Least Squares Fitter
(NLSF) (Fig 3).
To import the data, Origin's built-in pClamp import routine was
used.
|
Figure 1: Origin's pClamp Options dialog
|
Then, the data was plotted as a function of pre-pulse duration
(Fig. 2a), pre-pulse potential (Fig. 2b), recovery period (Fig.
2c). This showed the response of the ion channels during the pClamp
experiment.
|
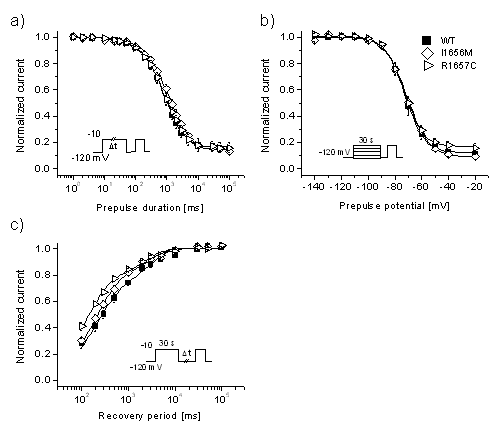
Figure 2 (a, b, c): This figure, a three-panel graph,
shows the electrophysiological response of normal (wild
type WT) and mutant (I1656M and R1656C) human neuronal voltage-gated
Na+ channel SCN1A (Nav1.1).
|
But characterizing the model graphically is only
qualitative. Implicit parametric information had to be extracted.
This was done using nonlinear regression analysis. Each of the
three sets of data was fit using the appropriate fitting model
in Origin's NLSF.
|
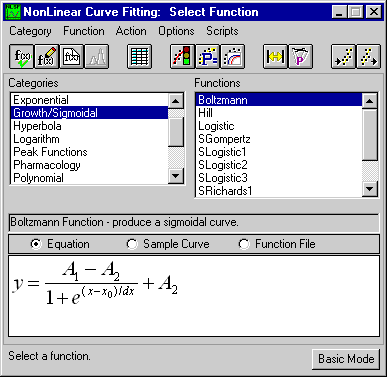
Figure 3: Origin's NLSF depicting a generic version
of the Boltzmann fitting model
|
- Data from onset of slow inactivation experiments was plotted
versus the length of the conditioning pulse and fitted to a second
order exponential decay function:
.gif)
where Ai describes the fraction of the
channels entering slow inactivation with time constant τi,
and y0 describes fraction of the channels
that remains active.
- Steady-state slow inactivation data was fitted to the Boltzmann
function:
.gif)
where V1/2 is the voltage where half-maximal
slow inactivation occurs, k is the slope factor
of the fit, and y0 is the fraction of
the channels that remains active.
- Recovery from slow inactivation was fitted to the second order
exponential function (referred to as the Exponential Associate
function in Origin):
.gif)
where Ai refers the fraction of the channels
recovering with time constant τi.
The NLSF interface allows complete control over the fitting process and is used to generate the published fit curves above (Fig.2) and the published parameters that clarify the Na+ channel functions. Results are presented as means ± standard error.
Acknowledgements
This work is part of a series of experiments reported in J Neurosci. 2003 Dec 10;23(36):11289-95.