Automating pKa Curve Fitting Using Origin
Summary
|
Brian Bissett, a scientist at Pfizer Global Research &
Development, has performed research related to the development
of new medicinal drugs. His research involved the analysis
of chemical compounds in order to determine their pKa values.
These pKa values were then, in turn, used to predict whether
or not a specific compound has or does not have drug-like
properties.
|
|
|
The Problem
Calculating the pKa values was an arduous task.
Absorbance vs. pH data was captured on a scanning plate reader
spectrophotometer. A typical assay utilized a standard 96 well microplate
column-wise, resulting in an analysis of 12 compounds per plate,
each with 8 data points. Each row of the microplate had a different
value pH buffer in it (typically from pH 3.0 - pH 10.0). A set amount
of compound in Dimethyl Sulfoxide (DMSO), a common pharmaceutical
industry solvent, was then dispensed down an entire column providing
absorbance values across the entire pH range for each compound.
A pKa could be determined provided the compound for analysis had
a perturbable pH induced chromophore*.
The real bottleneck was the fact that it took so much time to individually
curve fit each dataset to obtain a resultant pKa. First the scientist
had to choose "the best" wavelength from which to try
and curve fit a pKa. (The particular spectrophotometer being used
scanned from 250nm to 750nm in 5nm increments.) Once the optimal
wavelength was determined, the scientist would specify initial conditions
(usually "reasonable" guesses for a parameter's starting
value which would be optimized during the fitting process) for a
curve fit. When a fit was then obtained from a set of data, a decision
had to be made as to whether the fitted curve was returning a valid
result.
How Origin Was Used
To circumvent these difficulties and free up time, algorithms
were written in LabTalk™, Origin's scripting language, to
accomplish the following tasks:
-
determine the best wavelength from which to
extract data for a pKa curve fit
-
set up the curve fitter with reasonable initial
conditions and trigger a curve fit of the data at the chosen
wavelength
-
determine if the pKa returned from the fitting
session was indeed reasonable
The scripts were then hooked up to a graphical user interface,
or GUI. Figures 1 and 2 depict the tool for an early (circa 1995)
pKa assay developed by Mr. Bissett and a colleague, Dr. Chris Lipinski,
in what was then the Physical Measurements Laboratory at Pfizer.
|
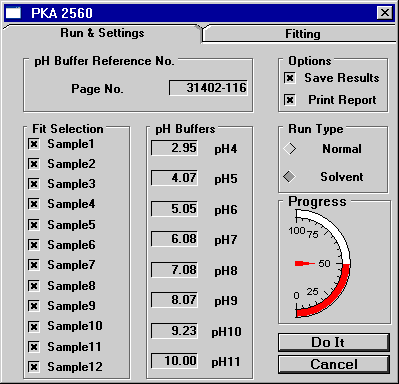
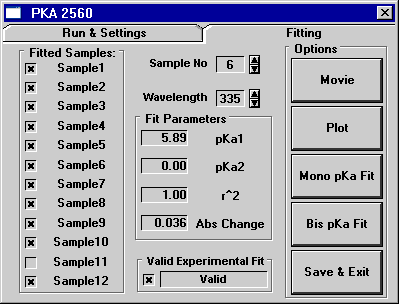
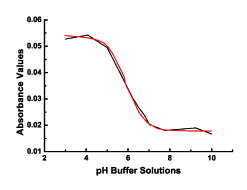
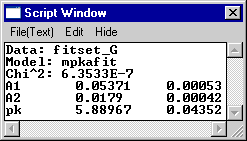
Figure 1: A solved single (or monotonic) pKa system
utilizing Origin.
|
The figure above shows the two tabs of the tool designed in Origin
for analyzing the pKa data. In this example, a monotonic (or single)
pKa system is solved. Pressing the "Do It" button in the
lower right corner of the Run & Settings tab caused the software
to automatically fit the data for all 12 samples using a rules based
system. It was also possible for the operator to hand fit the data
to any wavelength using either the bis or mono pKa equation by using
the Fitting tab. The fitted pKa's are shown as well as the r^2 (or
cod) value (which represents the "goodness" of the fit).
This option was typically only utilized if the automatic curve fitting
algorithms were incapable of settling on a solution. The plot shows
the raw data points in black with the fitted curve in red.
Below (Fig 2) is the identical screen but run on a dual (or bis)
pKa system.
|
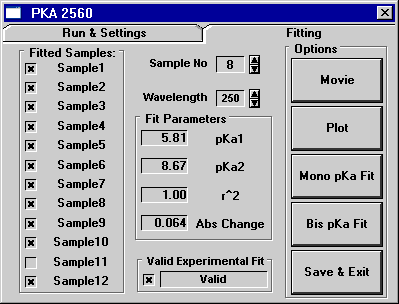
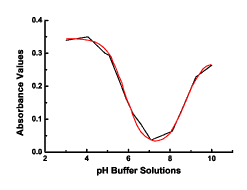
Figure 2: A solved bis (or two) pKa system using
Origin. Note: The Run & Settings tab is the same,
so it is only shown in Figure 1.
|
Notice that on the Fitting tab there are five buttons on the right
side. The top-most button, titled "Movie", allows the
user to view time lapsed screen shots of the absorbance vs. pH Buffer
Solution values as they change across the captured wavelength spectrum
in the plot window. This is an excellent tool for allowing a scientist
to discover which wavelength or series of wavelengths would best
lend itself to analysis.
Once the proper wavelength has been chosen, the data series can
then be plotted by pushing the "Plot" button. The wavelength
specified in the spin box control will determine which data series
or wavelength (250nm - 750nm) will be plotted, and the sample specified
in the sample spin box will determine which sample (1-12) will be
viewed or plotted. Once a sample has been plotted it can be fit
to either the mono or bis pKa equations by pushing either button.
If the user is satisfied with the resultant pKa returned, the results
can be saved, along with the compound's structure, to a database
file by pressing the "Save & Exit" button.
Biography
Brian Bissett
is a scientist in the Molecular Properties Group at Pfizer Global
Research & Development in Groton, CT. He has written a book
"Practical
Pharmaceutical Laboratory Automation", which has an entire
section dedicated to automating data analysis tasks utilizing Origin.
He also maintains a companion web site for the text at this location:
http://www.pharmalabauto.com/.