Properties of mPSCs at the C. Elegans Neuromuscular Junction
Analyses of mPSCs may be used to address a variety of important questions of synaptic release. For example, a change of mPSC frequency without a change of mean mPSCs amplitude suggests that presynaptic release probability may be changed. A change of mean mPSC amplitude suggests that the quantal size (amount of neurotransmitters released from exocytosis of a single synaptic vesicle) or postsynaptic receptor sensitivity may be changed. A single peak bell-shaped (normal) distribution in mPSC amplitude histogram suggests that mPSCs are probably due to exocytosis of a uniform population of synaptic vesicles. A positive correlation between mPSC amplitude and 10-90% rise time suggest that large-amplitude mPSCs might be due to synchronized multivesicular release. In combination with measurement of evoked postsynaptic currents (ePSCs), mPSCs may be used to estimate the quantal content (number of synaptic vesicles released per ePSC). |
|
click for larger image
|
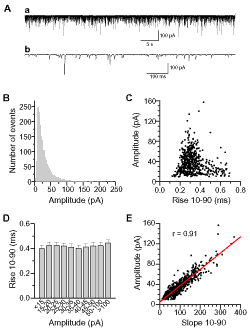
This example shows the properties of miniature postsynaptic currents (mPSCs) at the C. elegans neuromuscular junction.
In the absence of action potentials or direct nerve stimulation, synaptic vesicles at the presynaptic site may release neurotransmitters spontaneously, causing mPSCs in the postsynaptic cell membrane. At the C. elegans neuromuscular junction, mPSC amplitudes show great variability with many large-amplitude events. The analyses were performed to determine whether large-amplitude mPSCs were due to synchronized release of multiple synaptic vesicles.
The graph consists of several layers. There are three ways of making this type of graph using Origin: (1) make separate graphs and organize them in a Layout page, (2) make multiple graphs as separate layers in the same page by adding layers to the graph window, and (3) create separate graph windows and merge them into one graph window. The second approach was used in this example.
Figure A(a).
A segment (1 min) of an mPSC recording trace is shown in Figure A(a).
Importing the data. To make the graph, data acquired with the pClamp was imported into Origin. First, the pClamp Import dialog was opened by selecting File: Import: pClamp. In the dialog, the Files of type drop-down list was set to *.abf (the alternative choice *.dat is for data files acquired with older versions of pClamp). The desired trace file was then selected and added by clicking the Add file(s) button. After clicking OK, a pClamp Option dialog appears. The Channels option was selected and the file imported by clicking the Import button.
The imported data is displayed in a worksheet in Origin with a column label of Ch0(Y) pA, where Ch0 indicates the input channel from which the pClamp data was acquired, Y indicates that values in the column are of the y-axis type in an x-y plot, and pA is the unit of the data shown in the column. To reveal the X values, the View: Show X Column menu was selected. Note: This step is unnecessary since the X column is automatically used for the x-axis values during plotting even when it is not shown.
Plotting the graph. Column Ch0(Y) was selected by clicking on the column title and plotted as a Line plot by selecting Plot: Line. The orientation of the page was changed to Portrait using Page Setup (File: Page Setup). Control over text labels in the graph was then updated to a Fixed Factor of 1 in Plot Details by selecting Format: Layer and updating the Display tab. While still in the Plot Details dialog box, the layer dimensions were updated on the Size/Speed tab. The X and Y axes and tick labels were disabled on the Title & Format and Ticks Labels tabs, respectively, in the X and Y Axis dialog boxes. However, it is also possible to disable these features by de-selecting the X and Y Axes check boxes on the Display tab in Plot Details at the Layer level. Finally, a new XY scaler object was added by selecting Graph: New XY Scaler.
Figure A(b).
Figure A(b) represents a portion of the trace shown in Graph (A, a) but at an expanded time scale.
Creating the Graph. To create Figure A(b), Figure A(a) was copied and pasted onto the same graph page using the Edit: Copy Page and Edit: Paste menu commands. This creates an identical graph but in a new layer. This new layer was then linked to Figure A(a) by selecting Format: Layer to display Plot Details and then selecting Layer 1 from the Link to drop-down list on the Link Axes Scales tab. Control over the sizing of text labels was then updated to a Fixed Factor of 1 on the Display tab. Finally, the measurements for the layer were updated to be the same size as Figure A(a) (i.e. the parent layer) by selecting % of Linked Layer from the Units drop-down and entering 100 (percent) for the Width and Height fields on the Size/Speed tab. The x-axis range was then updated to go from 11000 to 12000 ms by modifying the From and To settings in the XAxis dialog box (Format: Axes: X Axis => Scale tab). To complete the Figure, the XY scaler was resized to x = 100 ms and y = 100 pA.
Figure B.
Figure B is a Column/Bar plot that shows amplitude distribution of mPSCs. Averaged mPSC amplitude distribution of 25 experiments is shown as Number of Events/min using a bin-width of 2 pA. MiniAnalysis (Synaptosoft, Decatur, GA) was used to analyze mPSCs, and the results were pasted into an Origin worksheet.
Creating the Graph. The Layer tool (Tools: Layer) was used to create a new layer in the graph that now contains Figures A, a and A, b. Note: On the Add tab in the Layer tool, the first toolbar button was clicked. This option adds a new, unlinked layer to the graph. After adding the layer, the position and size of the layer was fine tuned by editing the Size/Speed tab at the Layer3 level in Plot Details (Format: Layer). As with the previous layers, control over the sizing of text labels was updated to a Fixed Factor equal to 1 on the Display tab. Finally, the data was added to the layer as a Column/Bar plot using the Plot Setup dialog (Graph: Plot Setup).
Figure C to E.
Figures C and E are scatter plots showing correlation between mPSC amplitude and rise time (C), and between mPSC amplitude and rise slope (E), while Figure D shows the rise times of mPSCs grouped according to their amplitudes as a Column/Bar plot. Properties of mPSCs were analyzed with MiniAnalysis, and the results were copied into Origin worksheets.
Creating Each Graph. A new layer is created in the graph for each figure using the Layer tool as above. To maintain the relative sizes among graphs, the layers representing Figures C through E are linked to the layer representing Figure B by editing the settings at the layer level in Plot Details. In Figure E, the Fit Linear function of Origin was used to obtain the linear fit and the r value.
Acknowledgements
This figure was made by Qiang Liu and Zhao-Wen Wang. Additional examples made with Origin may be found in the following two publications:
Related: http://www.jneurosci.org/cgi/content/full/25/29/6745
Other: http://www.jbc.org/cgi/content/full/281/12/7881